
DYOGEN group
web-code version: 2014-09-19
database version: 89.01
 Contact us.
Contact us.

|

|

|
|
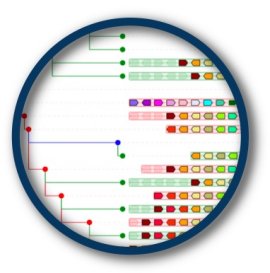
| Root species for fgf1a (ENSDARG00000017542) (Zebrafish) | AlignView depth | PhyloView depth | |
|---|---|---|---|
| Fungi/Metazoa group |  ~1500 Mya ~1500 Mya |
130 species  |
|
| Bilateria |  ~580 Mya ~580 Mya |
128 species  |
274 homologs (oldest homol.)  |
| Olfactores |  ~550 Mya ~550 Mya |
124 species  |
270 homologs  |
| Craniata |  ~550 Mya ~550 Mya |
120 species  |
267 homologs (dupl. node)  244 homologs (dupl. node)  137 homologs  |
| Euteleostomi |  ~420 Mya ~420 Mya |
118 species  |
136 homologs  |
| Neopterygii |  ~330 Mya ~330 Mya |
21 species  |
39 homologs  |
| Clupeocephala |  ~300 Mya ~300 Mya |
19 species  |
37 homologs (dupl. node)  19 homologs  |
| Otophysi |  ~150 Mya ~150 Mya |
3 species  |
3 homologs  |
| Zebrafish |  ~0 Mya ~0 Mya |
1 species  |
1 homolog  |
-evalue <Real>
Expectation value (E) threshold for saving hits
Default = '10'
-word_size <Integer, >=2>
Word size for wordfinder algorithm
Default = '3'
-gapopen <Integer>
Cost to open a gap
Default = '11'
-gapextend <Integer>
Cost to extend a gap
Default = '1'
-matrix <String>
Scoring matrix name (normally BLOSUM62)
-threshold <Real, >=0>
Minimum word score such that the word is added to the BLAST lookup table
Default = '11'
-window_size <Integer, >=0>
Multiple hits window size, use 0 to specify 1-hit algorithm
Default = '40'